The surprising properties of the coronavirus envelope
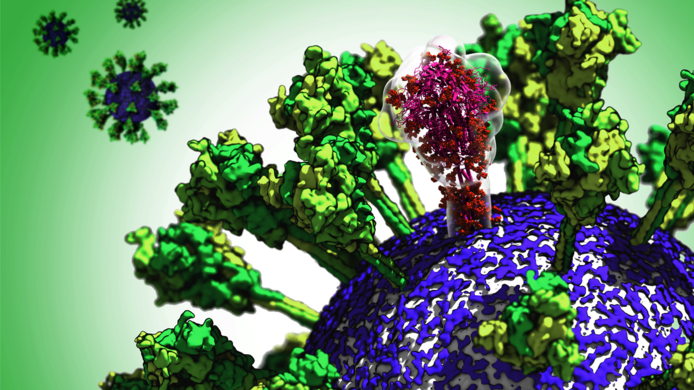
FWF: Mr. Sikora, you are an Erwin Schrödinger Fellow doing research at the Max Planck Institute of Biophysics in Frankfurt. What was the original subject of your project? Mateusz Sikora: I explored how living cells are bound together. There are certain proteins in the cell membranes that are like hooks binding cells together. This process is not well understood and it is not clear what triggers it, i.e. why some types of bonding are very strong and others are not. When the pandemic started, I soon realised that I had available exactly the right tools to study how the coronavirus binds to the cell, since viruses also use special proteins to attach themselves to the host cell – a prerequisite for infecting the cell. FWF: How did the pandemic change your focus? Sikora: I remember that back in February, when the pandemic hit Italy, we started talking in the corridor here at the Institute, wondering if anything was known about the surface of the virus. We thought that perhaps we could help, and I started to organise a task force on the coronavirus together with some other people, Professor Gerhard Hummer and my colleagues Sören von Bülow, Florian Blanc, Michael Gecht and Roberto Covino. The surface is important because it is the only part of the virus that is visible from the outside and therefore the only point of contact for the immune system or for drugs. The virus has a surface protein we call spike, which is shaped roughly like a lollipop, which it uses to attach itself to the host cell and initiate the connection between the cell membrane of the virus and that of the host cell.
We learned that some research groups had already succeeded in deciphering parts of the protein's structure. Its top part was already known in good resolution, but not how it is connected to the virus. They also did not know how well this image corresponded to “in-vivo” reality. Against this background, we started to develop a model of the protein and used all the facts we could find about the older coronavirus SARS-CoV-1. For these simulations we were accorded 20 million hours of computing time on the SuperMUC supercomputer in Germany. In addition, the FWF approved an extension of my fellowship, specifically for my work on Covid-19, so I was able to devote another four months to this work.
FWF: You were saying you were working with computer simulations. But the project also used images generated by electron microscopy. Sikora: That’s right, that was a nice synergy. At the same time as we were developing the models, Martin Beck's group here at the Institute had just received virus samples from Munich and prepared them for microscopic examination. They were working independently of us until they started to combine several images into a single high-resolution image of the spike protein. They found that they could not resolve the protein's stalk that connects it to the virus. They then approached us, and we already knew that this stalk was very flexible and elastic and were thus able to explain why they could not see this structure in their images. FWF: What is so unusual about this combination? Sikora: When we started our model calculations, we quickly saw that the head of the spike was very stable, but that the stalk had several hinges where it could bend. There was one hinge that worked like a knee joint, one that was like an ankle and one that was like a hip joint. We then started calling it a “leg” because the analogy was so compelling. FWF: What does this mean for the function of the protein? Sikora: You have to imagine that the virus is relatively small compared to the host cell, which is almost a thousand times larger. That means the surface of the host cell is practically a flat surface for the virus. If the spike was just a solid tip on the virus surface, only a single protein could bind to the host cell. The idea is that the coronavirus has flexible legs, so that several proteins touch the host cell at once. This increases the chances that a connection will actually be made.
FWF: What consequences do your findings have for research, especially for the development of vaccines? Sikora: Various companies and research groups have already been in touch with us. For them, the good news is that the in-vivo situation is very similar to what they had found in-vitro in the laboratories. They can trust in what has so far become known about the structure of the virus. Apart from that, the leg of the spike protein could be a good starting point for a drug or vaccine. If you could find an antibody that could bind to the leg, it would probably be very difficult for the virus to mutate in a way that would prevent this from happening. Also, the leg has changed very little between SARS-Cov-2 and its predecessor. So it seems to be an important part that allows the virus to function. FWF: Your results attracted a great deal of international attention. What does this discovery mean for you as a young researcher? Sikora: I was fortunate in that I had already developed the tools for this project during my Schrödinger Fellowship and that the extension of my research stay was approved. It is important for me now not to rest on my laurels. We are currently preparing several new publications. One of them centres on the results of a large simulation, with which we used to explore, among other things, which parts of the spike protein are so-called epitopes that form points of attachment for the immune system and can also be used as reference points for the development of vaccines.
Mateusz Sikora is a physicist who is currently engaged in research at the Max Planck Institute of Biophysics in Frankfurt through an Erwin Schrödinger Fellowship from the Austrian Science Fund FWF. Prior to this, he was a postdoc researcher at the Institute of Science and Technology (IST) Austria. He is interested in the modelling of biological molecules, especially the interaction of proteins embedded in biological membranes.
International Mobility
The Erwin Schrödinger Fellowships are a programme of the Austrian Science Fund FWF aimed at early-career researchers in Austria who would like to gather experience abroad after completing their doctorate. The fellowship, which also includes return phase support back in Austria, gives them the opportunity to learn about new fields and methods. The duration of a fellowship is between ten months and two years.
Publication






